Genomic Basis of Alternative Reproductive Strategies in the Ruff
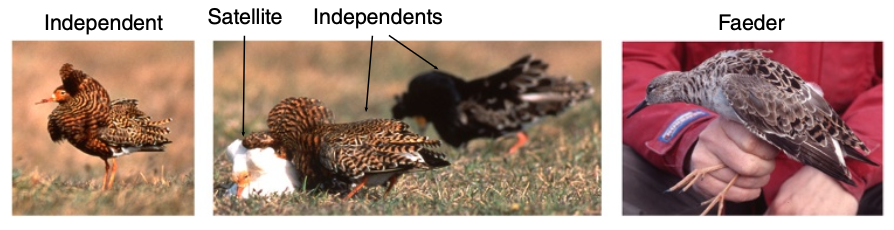
The ruff (Philomachus pugnax) is a migratory wading bird with one of the most complex and fascinating mating systems in the animal kingdom. Males exist in three distinct morphs—independents, satellites, and faeders—each differing in behavior, plumage, and body size. In our latest study, published in Nature Genetics, we investigate the genetic basis underlying these alternative reproductive strategies, revealing a 4.5-Mb inversion associated with dominant Satellite and Faeder morphs.
Key Findings
- High-Quality Genome Assembly
- We generated a 1.23 Gb genome assembly using Illumina sequencing with N50 scaffold size of 10 Mb.
- The ruff genome was compared against other birds to identify conserved synteny and evolutionary divergence.
- Discovery of a Large Chromosomal Inversion
- A 4.5-Mb inversion was detected on scaffold 28, perfectly correlating with Satellite and Faeder phenotypes.
- This inversion occurred ~3.8 million years ago, with a rare recombination event creating the Satellite chromosome ~500,000 years ago.
- Genetic Basis of Male Morphs
- Gene analysis identified missense mutations in MC1R, explaining plumage differences among morphs.
- Structural changes around HSD17B2 and SDR42E1, both involved in sex hormone metabolism, were linked to behavioral differences between morphs.
- Evolutionary Implications
- The accumulation of genetic changes within the inversion over time has resulted in a striking example of how structural variation drives phenotypic diversity.
- This work supports the broader concept of supergenes controlling complex traits, similar to findings in white-throated sparrows and butterfly mimicry genes.
Reflections
This research was conducted in collaboration with Professor Leif Andersson at Uppsala University and highlights how genomic analysis can uncover mutations tightly linked to behavioral differences in animals. While the ruff is well known in Europe, it has received limited cultural attention in China, making this study particularly unique in its scope.
Unlike plant genome research—where functional validation is often required—animal genome studies thrive on evolutionary storytelling. The ruff serves as a powerful example of how genetic variation shapes social behavior and reproductive strategies, reinforcing the importance of structural genomic changes in species adaptation.
The full text of this study can be accessed online at Nature Genetics.