Population Genomics of Japanese Quail: Insights into Genetic Diversity and Domestication
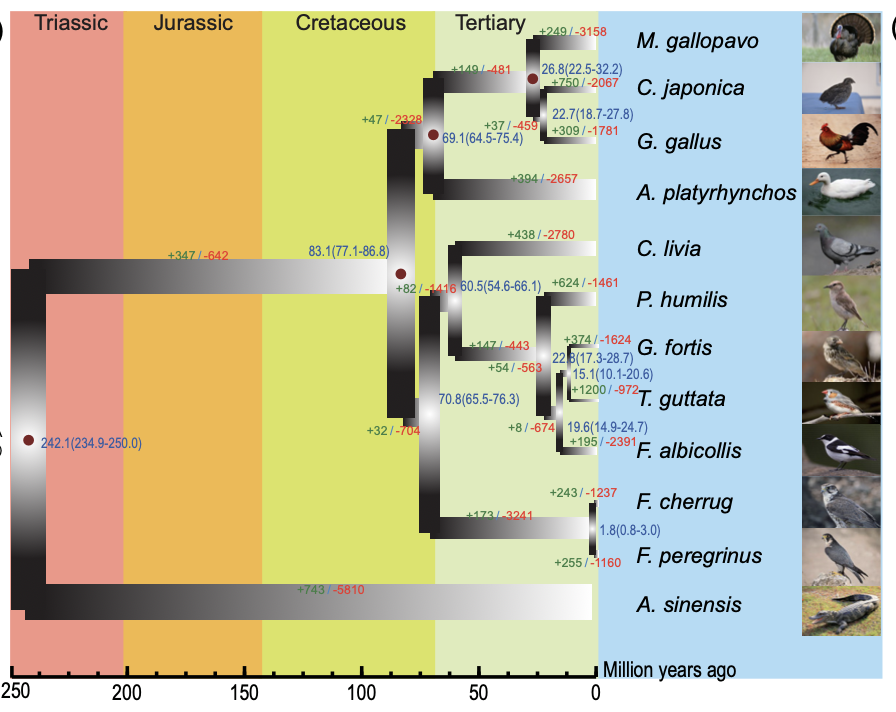
The Japanese quail (Coturnix japonica) is a poultry species with significant agricultural and scientific value. As a model organism for avian genetics, its rapid maturation and distinct genetic traits make it ideal for studying domestication and evolutionary processes. In our latest study, published in GigaScience, we present a chromosome-level genome assembly and population genomic analysis of Japanese quail, shedding light on genetic diversity, domestication history, and key traits related to breeding selection.
Key Findings
- High-Quality Genome Assembly:
- We generated a 1.04 Gb genome assembly, with 86.6% of sequences anchored to 30 chromosomes (28 autosomes and 2 sex chromosomes Z/W).
- Comparative analysis with chicken and turkey genomes revealed phylogenetic relationships within the Phasianidae family, clarifying a long-standing debate over evolutionary divergence.
- Genetic Basis of Early Maturation:
- The study identified four genes under positive selection associated with rapid sexual maturation in quail, including FSHB, a hormone gene influencing reproductive timing.
- Comparative hormonal analysis showed higher levels of follicle-stimulating hormone beta (FSHB) in quail, correlating with their accelerated growth and reproductive cycle.
- Population Genomics and Selective Sweeps:
- Genome-wide resequencing of 31 quail samples (wild, egg-type, and meat-type) revealed distinct selective sweep regions in domesticated lines, supporting the hypothesis of independent selection processes for egg-laying and meat production traits.
- A 1.8 Mb sweep region on chromosome Z was linked to plumage color, providing a valuable genetic marker for breeding programs.
- Genome-Wide Association Study (GWAS) for Plumage Color:
- GWAS identified a haplotype block on chromosome Z strongly associated with maroon/yellow plumage variation in domesticated quail.
- The study pinpointed CCDC171 as a key candidate gene, diverging from the previously known TYRP1 pigmentation gene found in other avian species.
Reflections
This project was conducted in collaboration with the Hubei Academy of Agricultural Science, where our team primarily contributed to high-throughput sequencing and genomic data analysis. By leveraging large-scale sequencing techniques, we successfully assembled and analyzed the quail genome, uncovering important genetic patterns related to domestication, selection, and evolutionary processes. These findings provide essential resources for genetic improvement in quail breeding, with broader implications for poultry genomics.
The full text of this study can be accessed online at GigaScience.