Chromosome-Level Comparative Analysis of Brassica Genomes
Brassica species are essential for agriculture and evolutionary studies, with genomes shaped by polyploidization events and extensive structural variations. In our latest study, published in Plant Molecular Biology, we present a chromosome-length genome assembly of Brassica nigra and a comparative analysis of Brassica chromosomes, shedding light on evolutionary divergence and structural variations.
Key Findings
-
High-Quality Brassica nigra Genome Assembly:
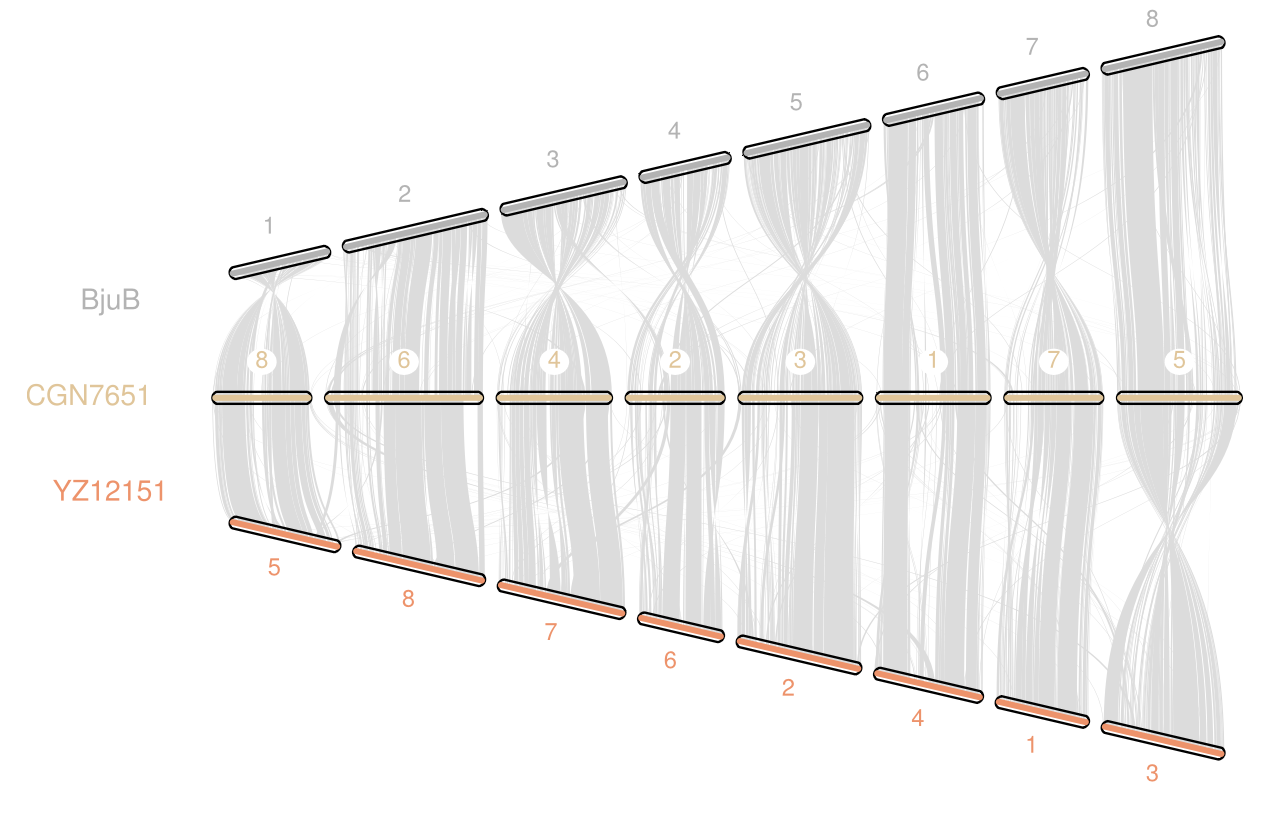
Using Hi-C sequencing, we generated an assembly of 484 Mb, with 393 Mb anchored onto 8 chromosomes, improving previous references with higher resolution and gene annotation accuracy. - Comparative Analysis of B Genomes in Mustard Species:
- The assembled B. nigra genome was compared with the B subgenome of B. juncea, revealing significant chromosomal variations post-polyploidization.
- Regions with major variations exhibited lower gene density and higher transposable element (TE) content, reinforcing the role of genome architecture in shaping species divergence.
- Evolutionary Insights into Brassica Chromosome Differentiation:
- Comparative chromosome mapping of Brassica A, B, and C genomes demonstrated that A genome remains closer to their common ancestor, while C genome underwent more structural variations after divergence.
- Identified orthologous regions across three Brassica diploids, refining our understanding of chromosome evolution and genomic plasticity in Brassica species.
Reflections
I first met Wenliang Wang when I joined BGI, and over the years, he has proven to be a dedicated expert in bioinformatics. This research formed the basis of his doctoral dissertation and was carried out in collaboration with our partners in Wuhan.
This project was initiated with a vision to leverage newly developed sequencing platforms for large-scale genome studies, and Brassica nigra—an essential but often underrepresented species—was selected for population genomic research. With its 10 Gb genome size, conducting resequencing and assembly posed significant computational challenges, requiring advanced scaffolding techniques such as Hi-C mapping. Overcoming these technical hurdles, we successfully refined genome assembly and evolutionary analysis, offering valuable resources for Brassica genomics and breeding applications.
The full text of this study can be accessed online at Plant Molecular Biology.