Molecular Digitization of a Botanical Garden: High-Depth Genome Sequencing of 689 Vascular Plant Species
Genome sequencing has transformed plant research, offering insights into species diversity, evolution, and adaptation. However, the number of plant species with fully sequenced genomes remains limited. In our latest study, published in GigaScience, we present high-depth whole-genome sequencing of 689 vascular plant species collected from Ruili Botanical Garden, China, marking an important step toward large-scale plant genome studies.
Key Findings
- Extensive Genomic Data Generation:
- We collected 1,093 samples, sequencing 761 plant specimens, generating a dataset of 54 Tb with an average sequencing depth of 60X per species.
- Voucher specimens were cataloged and stored in the Herbarium of China National GeneBank, ensuring precise taxonomic reference.
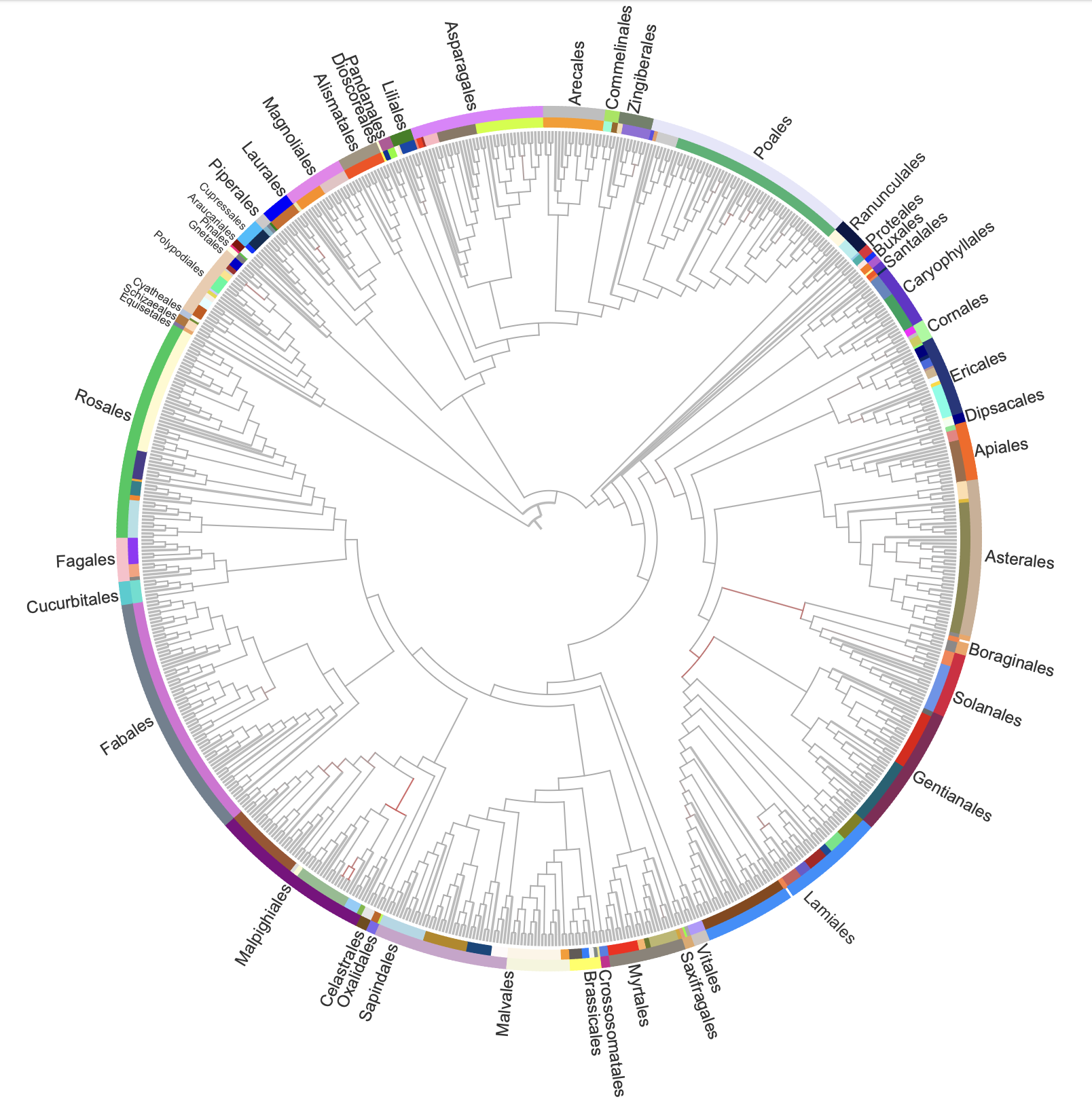
- Comprehensive Plant Identification Using Genomics:
- A total of 257 samples were identified at the species level, while 504 samples were classified at the family level, using chloroplast genomes and morphological analysis.
- We assembled chloroplast genomes for 689 species and constructed a phylogenetic reference tree using 78 chloroplast genes, enhancing species identification accuracy.
- Estimating Genomic Features:
- Using computational tools, we estimated genome sizes, repeat content, and heterozygosity for the sequenced plants, providing insights into genome complexity and structure.
- The dataset offers opportunities for studying plant genome evolution, polyploidy, and species divergence.
Reflections
After BGI established the National GeneBank, we introduced the concept of three core repositories: a sample bank, a database, and a living specimen collection. As part of our efforts to digitize biodiversity, we collaborated with botanical gardens, leveraging our expertise in genomic sequencing and data integration to initiate a series of plant genome studies. This project was conducted under this framework, with extensive sampling and sequencing at Ruili Botanical Garden.
Our mission was to enhance species identification, specimen preservation, and genome sequencing with a vision toward broader biodiversity conservation. Looking ahead, if we can continue refining genome assembly techniques, we will further improve plant genome research, enabling deeper evolutionary analyses that benefit the scientific community.
The full text of this study can be accessed online at GigaScience.