Resequencing 545 Ginkgo Genomes: Unveiling the Evolutionary History of a Living Fossil
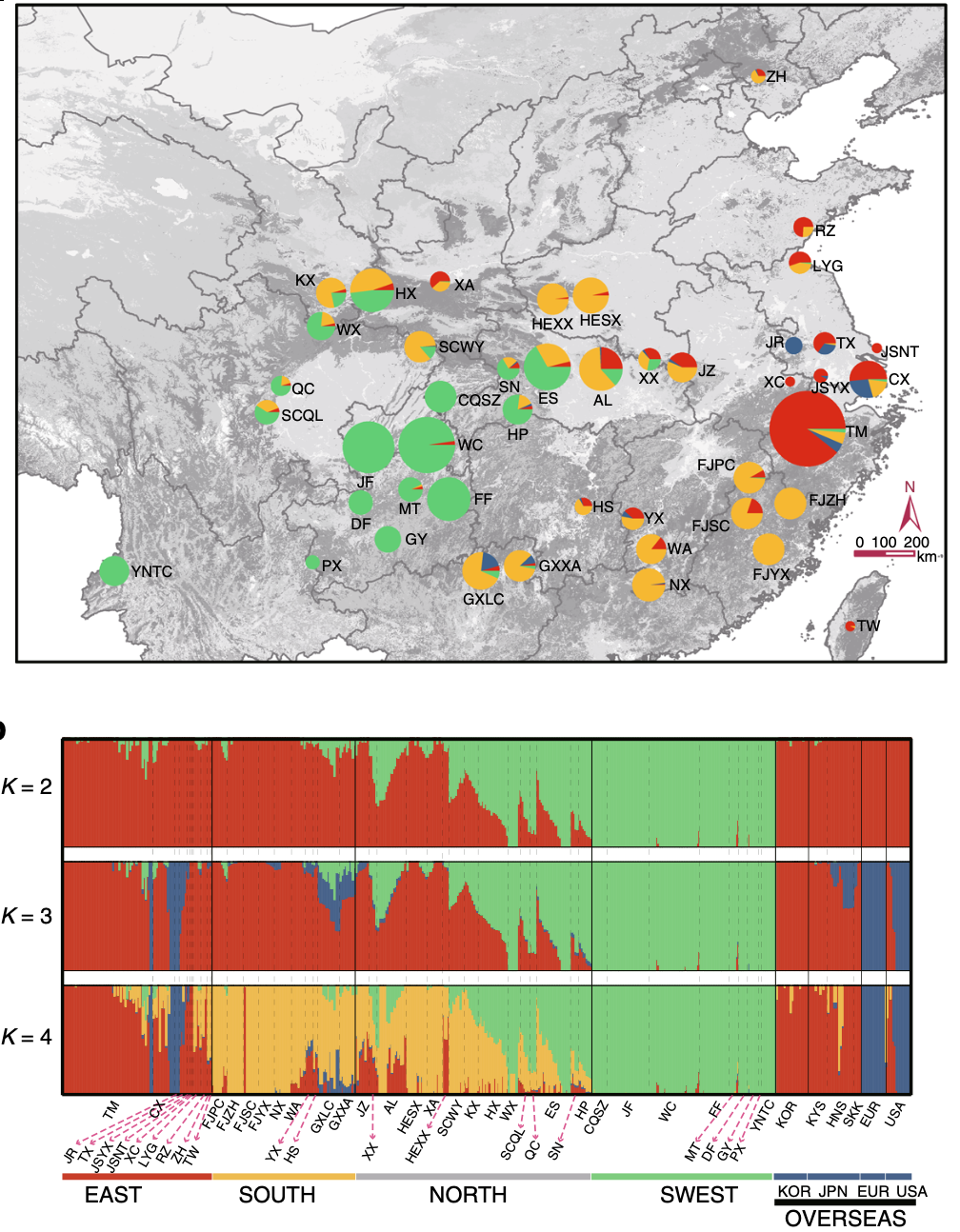
As one of the most ancient living plants, Ginkgo biloba has survived for over 200 million years, maintaining its resilience through dramatic environmental changes. In our latest study, published in Nature Communications, we present genome-wide resequencing of 545 ginkgo trees sampled across 51 global populations, offering unprecedented insights into ginkgo’s evolutionary history, population dynamics, and adaptations.
Key Findings
-
Identification of Three Refugia in China:
Our study confirmed that ginkgo populations persisted in three refugia—Southwestern, Southern, and Eastern China—during Pleistocene glaciations. Genetic analyses revealed four ancient genetic components, with populations in Northern China arising from admixture between Southern and Southwestern lineages. -
Anthropogenic Influence on Ginkgo Dispersal:
Phylogenetic and genetic analyses demonstrated multiple human-mediated introductions of ginkgo from Eastern China to Japan, Korea, Europe, and North America, supporting the hypothesis that people played a key role in the species’ global resurgence. -
Climatic Adaptation and Resilience:
Using species distribution modeling (SDM), we reconstructed ginkgo’s habitat shifts across historical climate events, showing northward expansion after the last glacial maximum (LGM). Bioclimatic factors such as temperature seasonality and precipitation patterns were identified as key drivers of ginkgo’s distribution. -
Natural Selection and Genetic Adaptation:
Genome-wide scans uncovered genes associated with stress tolerance, fungal resistance, and extreme weather adaptation, underscoring ginkgo’s ability to survive harsh environmental conditions.
Reflections
This project was initiated as part of a large-scale genome study leveraging our newly developed DNBSEQ sequencing platform. Professor Song Ge and Dean Xun Xu envisioned an ambitious effort to sequence high-quality plant genomes at scale, collaborating with Professor Yun-Peng Zhao from Zhejiang University, a leading expert in plant research. Given ginkgo’s historical, cultural, and scientific significance as a gymnosperm, we recognized the value of conducting a comprehensive population genomics study.
With a genome size of approximately 10 Gb, sequencing hundreds of individual trees posed massive data generation and analysis challenges. Overcoming these hurdles, we successfully completed the study, delivering valuable insights into ginkgo’s evolutionary and ecological dynamics. Beyond this publication, our extensive dataset allowed us to identify sex chromosomes, revealing an XY-based sex determination system in ginkgo. This finding is particularly exciting, as sex determination mechanisms in plants—especially ancient lineages like ginkgo—remain a fascinating subject for evolutionary biology.
Although our initial sex-determination study was not accepted alongside this work, we expanded our dataset by sequencing and assembling additional ginkgo genomes, refining our analysis of sex determination regions. We look forward to publishing these findings soon, further advancing our understanding of plant genetics and evolution.
The full text of this study can be accessed online at Nature Communications.