Draft Genome Sequence of Solanum Aethiopicum: Advancing Disease Resistance and Drought Tolerance Research
The African eggplant (Solanum aethiopicum) is a vital crop cultivated across tropical Africa for its nutritional and medicinal value. Despite its significance, limited genomic resources have slowed breeding progress compared to other Solanaceae relatives. In our recent study, published in GigaScience, we present a high-quality draft genome of S. aethiopicum, offering new insights into disease resistance, drought tolerance, and evolutionary mechanisms within orphan crops.
Key Findings
- High-Quality Genome Assembly and Annotation:
- We generated a 1.02 Gb draft genome, revealing a 78.9% repeat-rich structure.
- A total of 37,681 gene models were annotated, including 34,906 protein-coding genes.
- LTR Retrotransposon Amplifications in Disease Resistance Genes:
- Two major bursts of long terminal repeat (LTR) retrotransposon amplification (~1.25 and 3.5 million years ago) contributed to the expansion of disease resistance genes.
- These insertions facilitated resistance to pathogens like Fusarium, Ralstonia, and Verticillium, making S. aethiopicum an ideal genetic reservoir for crop improvement.
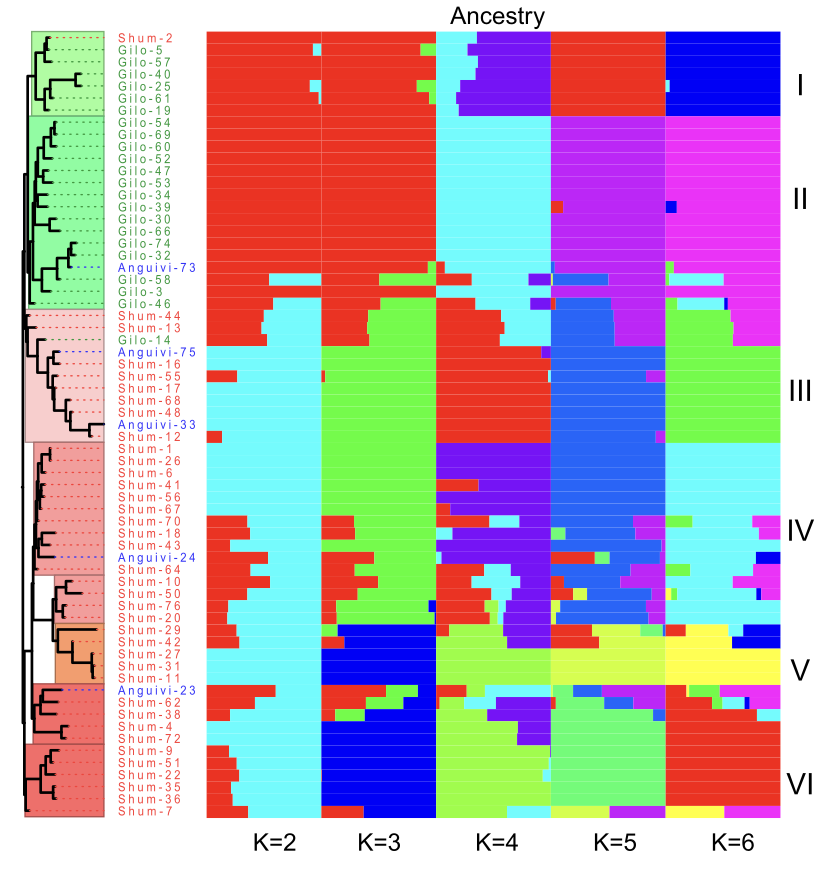
- Resequencing and Pan-Genome Construction:
- Genome-wide resequencing of 65 accessions (including S. anguivi, its wild ancestor) identified 18.6 million SNPs, with 34,171 SNPs linked to disease resistance genes.
- The pan-genome included 51,351 genes, uncovering 7,069 genes absent from the reference genome, broadening genetic resources for breeding initiatives.
- Domestication and Adaptive Evolution:
- Our study traced domestication patterns within the Gilo and Shum groups, identifying active selection in drought-resistance genes and environmental adaptation mechanisms.
- Population genetics revealed a bottleneck event ~4,000-5,000 years ago, followed by rapid population expansion, likely influenced by human cultivation.
Reflections
This research is part of the African Orphan Crop Consortium (AOCC) initiative, conducted in collaboration with UC Davis and ICRAF. The partnership provided essential sample resources, while our team led the genome sequencing and analysis. The project was efficiently driven by Bo Song, whose leadership ensured smooth execution and high-quality data production.
Following this successful study, Bo Song later transitioned to the Shenzhen Institute of Genomics at the Chinese Academy of Agricultural Sciences, where he continues his impactful work in plant genomics. This project exemplifies how advancing genomic research in orphan crops can enhance agricultural sustainability, improve breeding efficiency, and contribute to global food security.
The full text of this study can be accessed online at GigaScience.