Chromosome-Level Genome Assembly of Mangrove Fungus Penicillium variabile HXQ-H-1
Mangrove ecosystems host a diverse range of microorganisms, including fungi that play a crucial role in organic matter decomposition and secondary metabolite production. Among them, Penicillium variabile is a widespread ascomycetous fungus known for its ability to produce bioactive compounds with pharmaceutical potential. In our latest study, published in Journal of Fungi, we present the first chromosome-level genome assembly of P. variabile HXQ-H-1, a strain isolated from mangrove sediments along the coast of Fujian Province, China.
Key Findings
- High-Quality Genome Assembly:
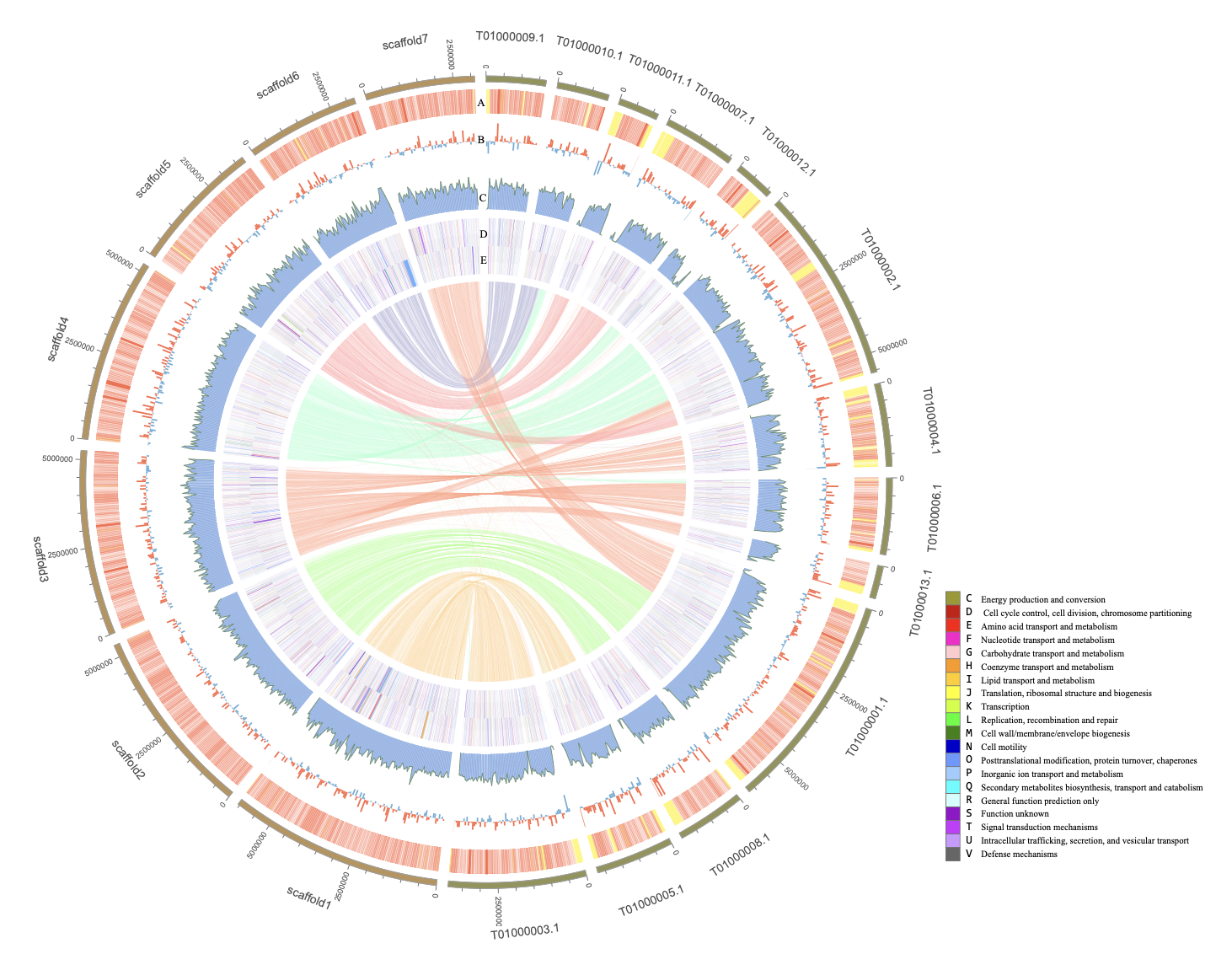
- The assembled genome spans 33.32 Mb, with a scaffold N50 of 5.23 Mb and contig N50 of 96.74 kb.
- Approximately 95.91% of the assembly sequences were anchored into seven longest scaffolds, providing a reliable genomic reference for further studies.
- Functional Genome Annotation:
- A total of 10,622 protein-coding genes were identified, with 99.66% of them annotated across eight functional databases.
- Key biosynthetic gene clusters involved in polyketide synthase (PKS), non-ribosomal peptide synthetase (NRPS), and terpene synthesis were found, highlighting the strain’s biotechnological potential.
- Comparative Genomics and Phylogenetics:
- Comparison with Talaromyces islandicus showed HXQ-H-1 has a slightly higher gene count, with enriched functional categories related to signal transduction, secondary metabolite biosynthesis, and energy metabolism.
- A genome-wide phylogenetic analysis confirmed that HXQ-H-1 is closely related to T. islandicus, further refining the classification of Penicillium and Talaromyces fungi.
Reflections
This research is part of our ongoing effort to explore fungal biodiversity in mangrove ecosystems, an area rich in untapped microbial diversity with significant ecological and biotechnological implications. The project was led by Ling Peng and Liangwei Li, with contributions from our broader research team, including Chengcheng Shi and Guangyi Fan.
This study represents an important milestone in fungal genomics, not only providing a high-resolution genomic reference for Penicillium variabile, but also paving the way for bioprospecting novel bioactive compounds and understanding fungal adaptations in mangrove environments. Moving forward, we aim to further investigate the functional roles of secondary metabolites and their potential applications in medicine and industry.
The full text of this study can be accessed online at Journal of Fungi.