Lineage-Specific Evolution of Mangrove Plastid Genomes
Mangrove forests thrive in coastal tropical and subtropical regions, bridging the gap between terrestrial and marine ecosystems. These plants have undergone independent evolutionary transitions from terrestrial ancestors, presenting an opportunity to explore genomic convergence and lineage-specific adaptations. In our latest study, published in The Plant Genome, we compared the plastid genomes of 21 mangrove species, revealing key evolutionary patterns and adaptations.
Key Findings
- Highly Conserved Plastid Genome Structure:
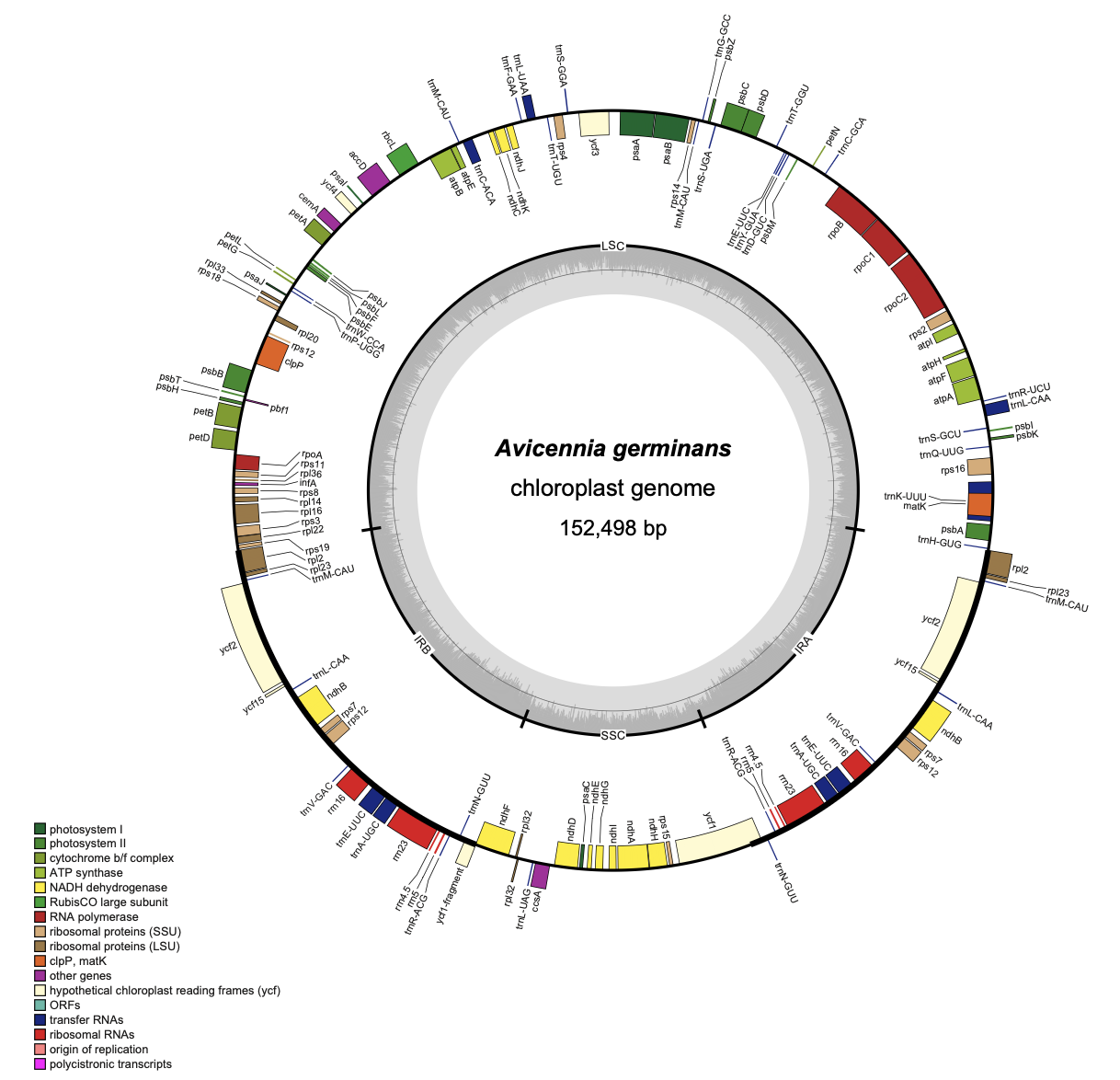
Despite their diverse evolutionary backgrounds, chloroplast gene order, content, and genome size remain largely conserved across mangrove species. However, specific deviations include:- Loss of the photosystem I gene (psaZ) in Acanthus ilicifolius
- Inversion of the ribosomal protein gene (rpl23) in Avicennia germinans
-
Phylogenetic Clustering and Repeat Content Variation:
Phylogenetic analysis grouped mangroves into six major clades: core leptosporangiates, magnoliids, commelinids, lamiids, malvids, and fabids. Repeat content analysis showed that repeats are conserved within species of the same order but vary across different mangrove lineages. - Positive Selection and Evolutionary Dynamics:
The ribosomal protein gene (rps7) was under positive selection in certain mangrove species, including Kandelia obovato, Rhizophora stylosa, Bruguiera sexangular, and Rhizophora mangle. However, no widespread signs of convergent evolution were detected in mangrove plastid genomes.
Reflections
This study is part of our broader research into mangrove symbiotic microbiomes, contributing valuable genomic resources for understanding mangrove adaptation and lineage-specific evolution. The research was led by Chengcheng Shi as part of their doctoral dissertation, with support from Xin Liu’s team at BGI-Qingdao.
Mangrove plants have evolved to withstand extreme environments, including high salinity, anaerobic soils, and tidal fluctuations. However, despite these similar ecological pressures, our study revealed minimal genomic convergence among chloroplast genomes. Instead, lineage-specific evolutionary dynamics appear to play a greater role in shaping plastid genome variations. As we continue our environmental genomics research, we aim to further explore how plastid evolution influences plant adaptation in extreme habitats.
The full text of this study can be accessed online at The Plant Genome.