Microbial Diversity in Industrial Effluents: Unlocking Source Tracking Potential
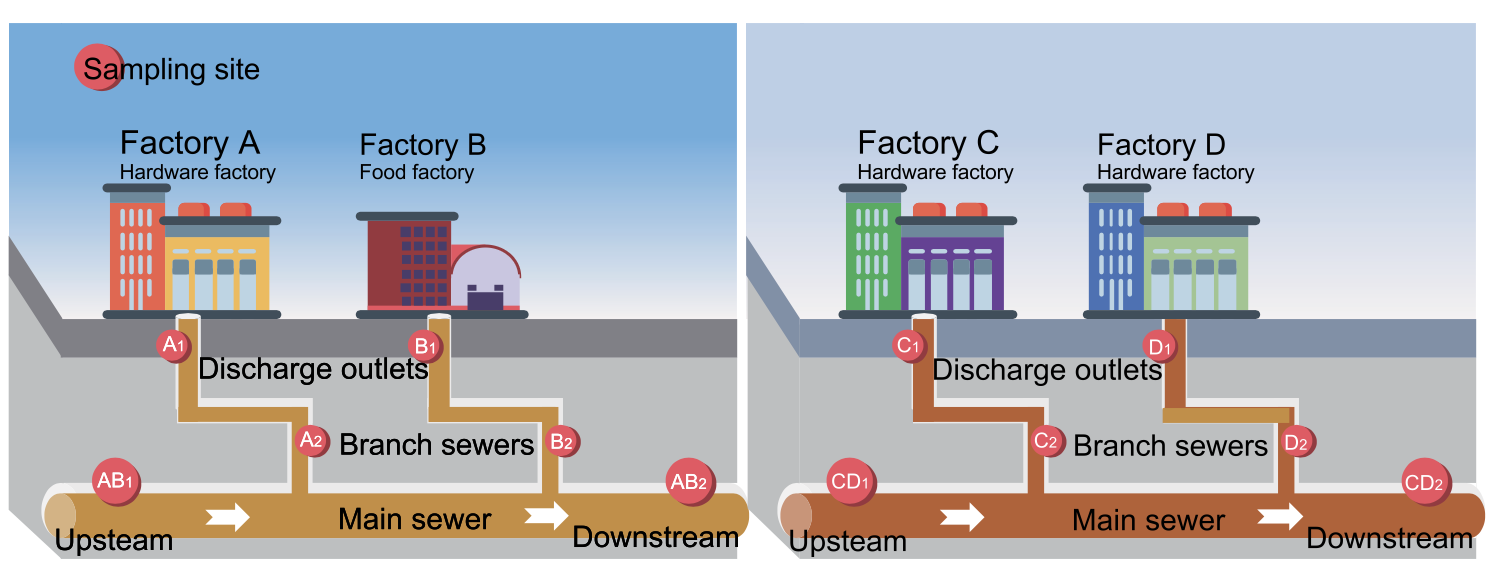
Understanding the microbial diversity in industrial effluents is crucial for efficient environmental surveillance and management. In our recent study published in Environmental Research, we explored microbial compositions within industrial sewage collected from factories in Shenzhen, China. Using high-throughput 16S rRNA gene sequencing, we identified microbial markers that enable source tracking of industrial effluent discharges. This innovative approach underscores the application of sequencing technology in environmental monitoring.
Key Findings
-
Microbial Diversity Across Sewer Networks:
We collected 28 sewage samples from different sewer locations connected to four factories and identified 5413 operational taxonomic units (OTUs) in total. While only 107 OTUs were shared by 90% of the samples, each factory’s effluents exhibited distinct microbial profiles, reflecting their unique industrial processes. -
Source Tracking Using Microbial Markers:
By comparing microbial compositions, we identified factory-specific microbial markers that differentiate effluents from the four factories. For instance, hardware factories were characterized by microbes capable of surviving in nutrient-limited environments and degrading heavy metals, while the food factory effluents contained microbes specialized in digesting organic matter. These microbial markers demonstrate the feasibility of using sequencing data for industrial effluent source tracking. -
Potential for Expanded Research:
This study not only highlights the promise of microbial source tracking but also opens avenues for more comprehensive research utilizing metagenomic data. Metagenomics could provide deeper insights into functional genes and microbial interactions, enabling broader applications in environmental genomics.
Reflections
This study represents our first foray into the field of environmental research. It originated from discussions with our collaborators in Shenzhen, who were exploring applications of genomic technology in environmental monitoring. I proposed the idea of leveraging emerging environmental DNA methods for this project, and we jointly designed a study focused on sampling and sequencing microbial communities in sewer networks. While we limited this effort to 16S rRNA gene sequencing rather than full metagenomics, the results were striking—clear microbial distinctions between different locations and effluent sources were observed.
Collaborating with Prof. Pengsong Li from Beijing Forestry University, we successfully completed this study. Although we have since designed a series of environmental microbiome research projects, their progress has been slower than expected. Nonetheless, I believe this study lays the groundwork for further explorations into microbial diversity in industrial effluents and the practical applications of genomic technologies in environmental science.
The full text of this study can be accessed online at Environmental Research.