Chromosome-Level Genome Assembly of Fall Armyworm Sheds Light on Its Invasion and Pesticide Resistance
Fall armyworm (Spodoptera frugiperda) is a highly destructive pest that has rapidly spread across multiple continents, causing significant damage to staple crops like maize, rice, sugarcane, and sorghum. In our latest study, published in Protein & Cell, we present a chromosome-level genome assembly of fall armyworm, offering key insights into its genetic diversity, invasion history, and resistance mechanisms to pesticides. These findings provide a foundation for better pest control strategies and biosecurity efforts.
Key Findings
-
High-Quality Genome Assembly Using stLFR and Hi-C Data:
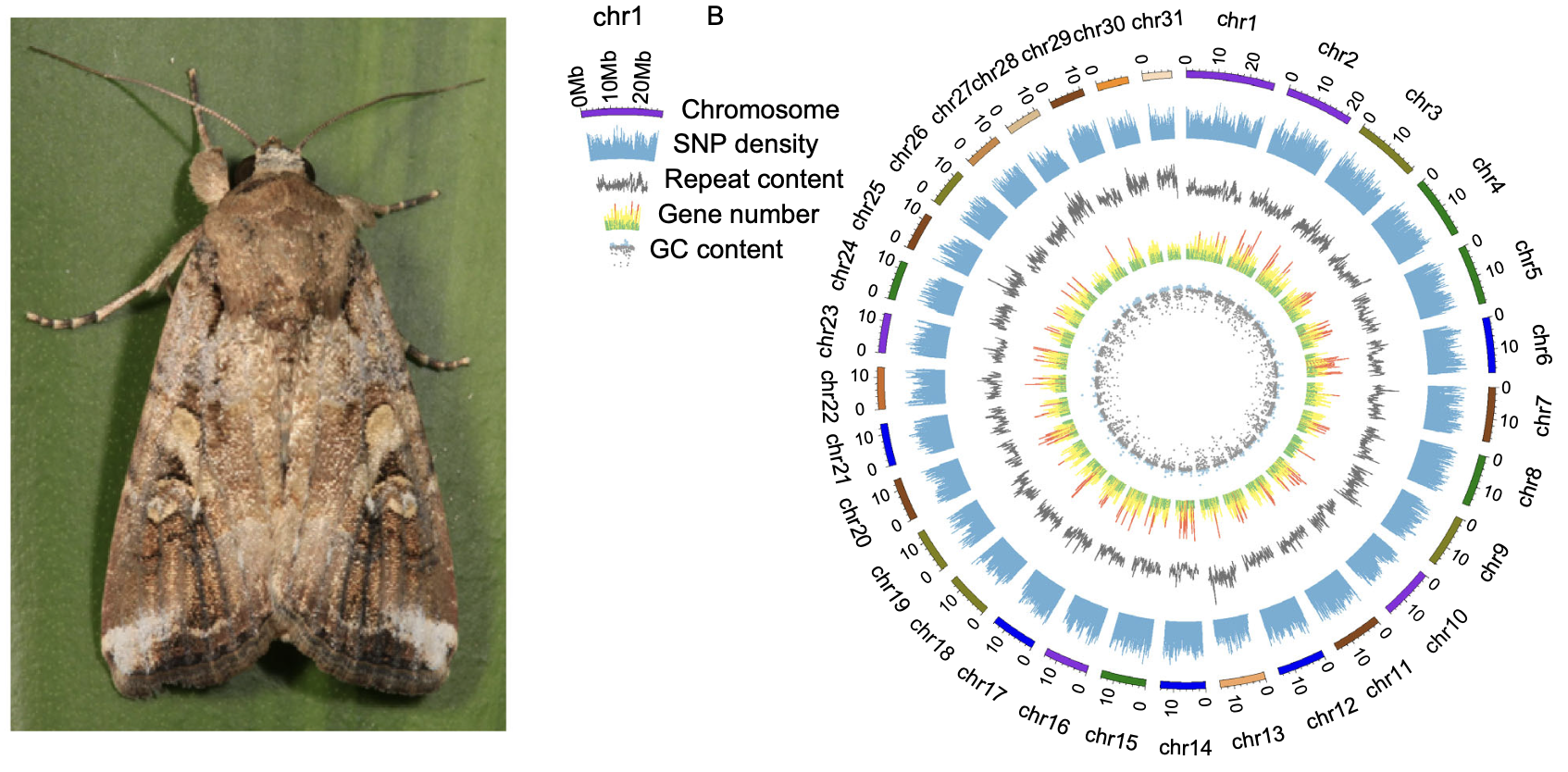
Our genome sequencing efforts resulted in a 542.42 Mb high-contiguity genome assembly, anchored to 31 chromosomes. Due to the complexity of insect genomes, assembling the fall armyworm genome was particularly challenging; however, the application of single-tube long fragment read (stLFR) sequencing alongside Hi-C scaffolding enabled us to produce a chromosome-level reference genome for the species. -
Origins and Population Evolution of Fall Armyworm in China:
Whole-genome re-sequencing of 163 individuals from America, Africa, and China revealed significant genetic differentiation between populations. Phylogenetic analysis confirmed that the fall armyworms invading China originated from African populations, rather than arriving directly from America. Additionally, strain identification showed that only the C strain was present in both Chinese and African populations, further supporting its migration pathway. -
Pesticide Resistance Mechanisms Identified Through Transcriptomics:
One of the most striking discoveries was the expansion of the cytochrome P450 gene family, which plays a crucial role in pesticide detoxification. The genome contains 425 P450 genes, of which 283 are unique to fall armyworm, making this an exceptionally large gene expansion compared to other lepidopteran species. Functional studies on gene expression in response to 23 different pesticides confirmed that several detoxification pathways—including AOX, UGT, and GST—were actively involved in resistance mechanisms.
Reflections
The year 2020 marked the beginning of the COVID-19 pandemic, but it was also a time when fall armyworm outbreaks became widespread in southern China. As part of our longstanding commitment to leveraging genomics for both public health and agricultural challenges, we saw an urgent need to decipher the genome of this invasive pest to mitigate its impact on crops.
Under the guidance of Professor Huanming Yang, we quickly established collaborations with researchers in Yunnan and Guangdong, securing the necessary samples for sequencing. Due to logistical constraints, we were unable to use conventional long-read sequencing methods, but our in-house stLFR technology proved highly effective for assembling the fall armyworm genome. Insects vary widely in genomic complexity, but for this species, our sequencing strategy delivered a robust genome assembly that enabled extensive downstream analyses.
Beyond genome assembly, our collaborators also pursued proteomic studies, adding further depth to our understanding of fall armyworm biology. The success of this research owes much to the efforts of Professor Yang Dong at Yunnan Agricultural University, whose contributions were instrumental in driving this project forward. Meanwhile, we are honored to collaborate with Academician Le Kang and his team. During my visit to his lab at Hebei University, I learned about their extensive and systematic research on locusts, which I found truly fascinating.
The full text of this study can be accessed online at Protein & Cell.