Chromosome-Level Genome of Butterflyfish Reveals Unique Color Patterns and Morphological Features
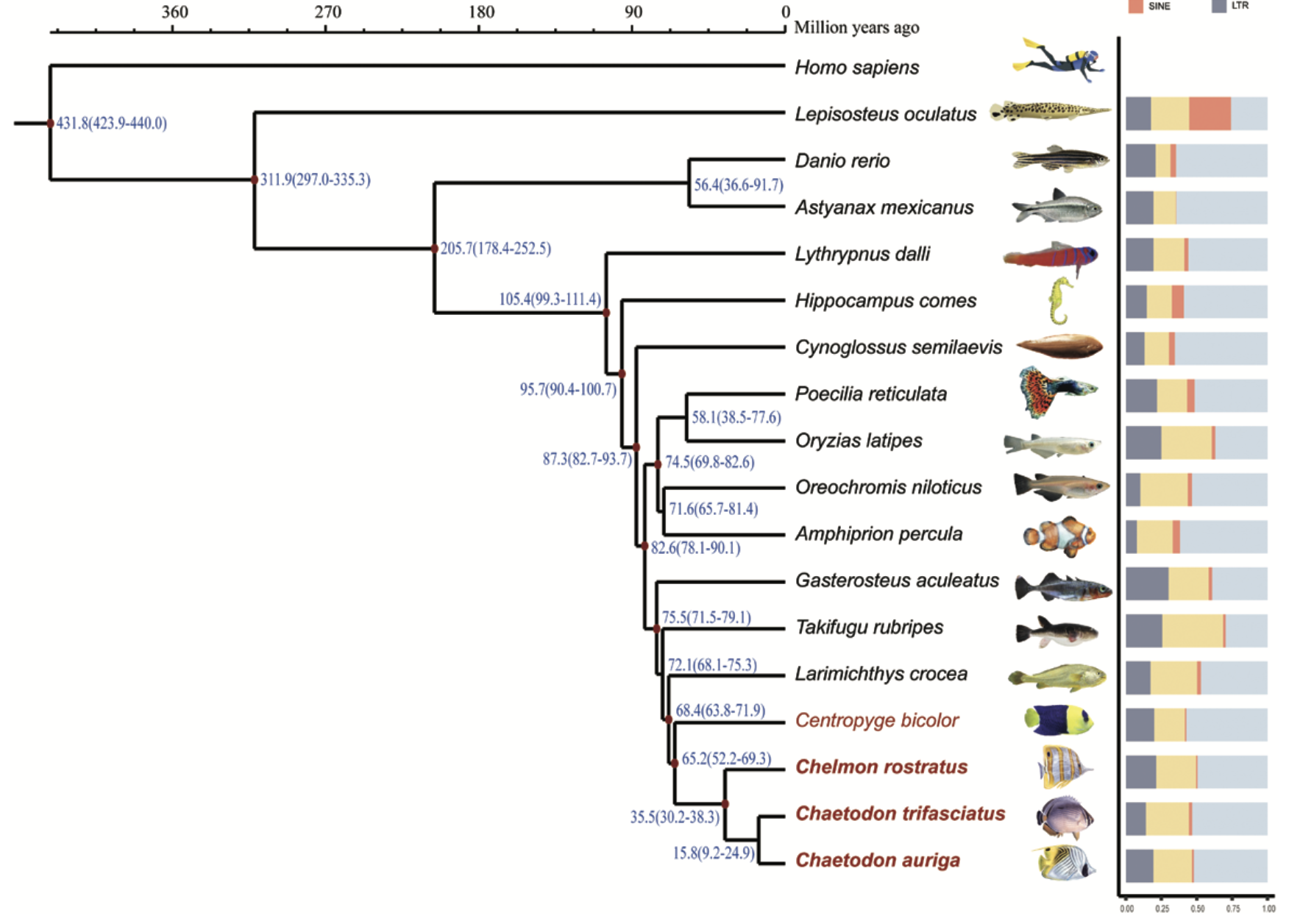
Butterflyfishes (family Chaetodontidae) are iconic reef dwellers, renowned for their vibrant color patterns and specialized jaw morphologies. Our recent study, published in DNA Research on August 17, 2023, provides the first chromosome-level genome assembly for butterflyfish (Chelmon rostratus) and additional genome assemblies for Chaetodon trifasciatus, Chaetodon auriga, and Centropyge bicolor (a closely related Pomacanthidae species). These genomic resources uncover the molecular basis behind the distinctive pigmentation and unique morphological adaptations in butterflyfish.
Key Findings
-
High-Quality Genome Assemblies:
The genome of Chelmon rostratus was assembled to the chromosome level, consisting of 24 pseudochromosomes with a total size of 638.70 Mb. This reference genome achieves a BUSCO completeness score of 97.3%, highlighting its quality. The other three genomes were also of high quality, providing a comprehensive resource for comparative analyses. -
A Novel Pigmentation Gene—tyrp3:
Our study identified tyrp3, a tyrosinase-related protein gene uniquely present in reef fishes like butterflyfish and clownfish. This gene, implicated in pigmentation diversity, may represent a reef-specific evolutionary adaptation for vibrant coloration. It adds to the previously known melanocyte-specific tyr genes (tyr1 and tyr2), offering new insights into fish pigmentation. -
Morphological Specialization through Gene Duplication:
In Chelmon rostratus, we observed an expansion of the nell1 gene (three tandem duplications), which is associated with jaw development and bone formation. This genetic adaptation likely underpins the species’ distinct long, sharp jaw, enabling its specialized feeding strategy on coral reef invertebrates.
These findings offer fresh perspectives on the genomic underpinnings of key traits in reef fishes and provide a foundational resource for further studies in fish pigmentation and functional morphology.
Reflections and Comments
Fish genome research is a major focus at the BGI-Qingdao Research Institute, where many members of XinLab are based, making fish genomics one of XinLab’s core research areas. This study was primarily led by my student, Zhang Suyu, who contributed significantly to the analysis of genomic data as part of our larger multi-species fish genome project. As a student at the University of Chinese Academy of Sciences, Zhang Suyu also needed to independently lead a project culminating in a first-author publication, which gave rise to this endeavor.
Zhang Suyu approached the project with dedication and responsibility, diligently conducting the bioinformatics and data analysis. His hard work ensured the smooth completion and successful publication of this study. This achievement underscores the collaborative and supportive spirit of our lab, as well as the rigor and passion our members bring to genomic research.
Conclusion
This research not only enriches the genomic resources available for butterflyfishes but also uncovers unique genetic features driving their vibrant coloration and specialized morphologies. By shedding light on these evolutionary adaptations, our study adds valuable knowledge to the field of reef fish genomics and opens new avenues for understanding fish pigmentation and ornamental traits.