Rattan Phylogeny based on the Calamus tetradactylus Chloroplast Genome
Our recent study published in BMC Genomic Data by Zhang et al. provides the first complete chloroplast genome sequence for Calamus tetradactylus, a valuable rattan species, shedding light on its genomic features and evolutionary relationships within the palm family (Arecaceae).
Background
- Calamus tetradactylus, commonly known as a small-diameter rattan, is primarily found in Vietnam, Laos, and southern China. It is highly valued commercially for its material properties.
- While its physical characteristics have been studied, its genomic information, crucial for understanding its evolution and aiding breeding programs, remained largely unexplored.
Methods & Findings
The researchers sequenced, assembled, and annotated the complete chloroplast (cp) genome of C. tetradactylus. Key findings include:
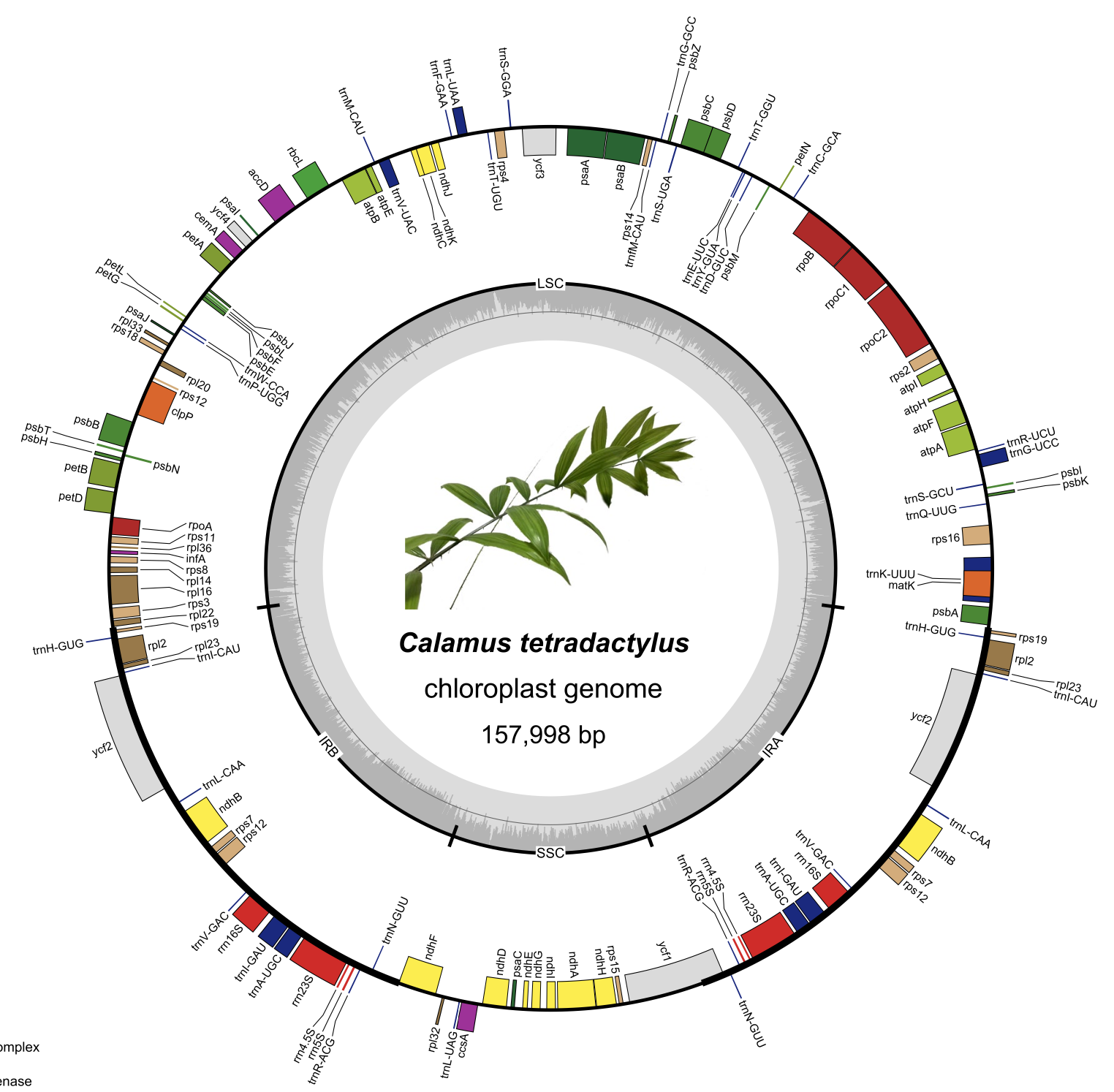
- Genome Structure:
- The cp genome is 157,998 bp long and displays the typical quadripartite structure found in most angiosperms: a Large Single-Copy (LSC) region (85,760 bp), a Small Single-Copy (SSC) region (17,602 bp), and two Inverted Repeat (IR) regions (27,318 bp each).
- It encodes 132 genes, including 86 protein-coding genes, 38 tRNA genes, and 8 rRNA genes.
- The overall GC content is 37.24%.
- Comparative Genomics & Variation:
- The cp genome structure is highly conserved compared to related Arecaceae species, though variations exist, particularly at the IR/SC boundary regions, contributing to genome size differences among species.
- Significant sequence divergence was found in non-coding regions, especially within the LSC and SSC regions, compared to the highly conserved IR regions.
- Specific genes (e.g.,
trnT-GGU,ycf1) and intergenic regions (e.g.,psbF-psbE,ndhF-rpl32) showed high nucleotide diversity (π). These variable regions are proposed as potential DNA markers for species identification and population genetics.
- Phylogenetic Analysis:
- Phylogenetic trees constructed using cp genome sequences placed C. tetradactylus firmly within the Arecaceae family in the Monocots clade.
- Within Arecaceae, C. tetradactylus forms a distinct clade with other Calamus species, confirming the monophyly of the genus.
- The analysis revealed that C. tetradactylus is most closely related to C. walkeri, aligning with previous morphological classifications.
Conclusion & Significance
This study provides the first complete chloroplast genome for C. tetradactylus, offering valuable genomic resources. The findings confirm its phylogenetic position, support the monophyly of the Calamus genus, and identify potential molecular markers for species identification. This genomic information is expected to benefit future rattan research, including evolutionary studies, breeding programs, and sustainable resource management.
Personal thinking
This study was conducted in collaboration with the International Bamboo and Rattan Center and supported by funding from the National Key R&D Program. The process, from obtaining samples and completing sequencing to defining the research goal of focusing on chloroplasts for fundamental analysis and interpreting evolutionary relationships, and subsequently completing the study, was accomplished in a relatively short time. It can be regarded as a highly efficient research process, with the submission and review process also being relatively smooth. Additionally, several genomes of bamboo and rattan species from this project will be analyzed in follow-up studies, with more related articles on bamboo and rattan genomics expected to be published. We hope that our team, especially Zhang Haibo and other colleagues involved, will continue to carry out the subsequent research work with the same level of efficiency.
Reference: Zhang, H., Liu, P., Zhang, Y. et al. Chloroplast genome of Calamus tetradactylus revealed rattan phylogeny. BMC Genomic Data 25, 34 (2024). https://doi.org/10.1186/s12863-024-01222-0