Cicer Super-Pangenome Study Offers Insights for Chickpea Improvement
Published online in Nature Genetics on May 23, 2024, a study presents genomes of eight wild chickpea relatives and a Cicer super-pangenome, providing resources for crop enhancement.
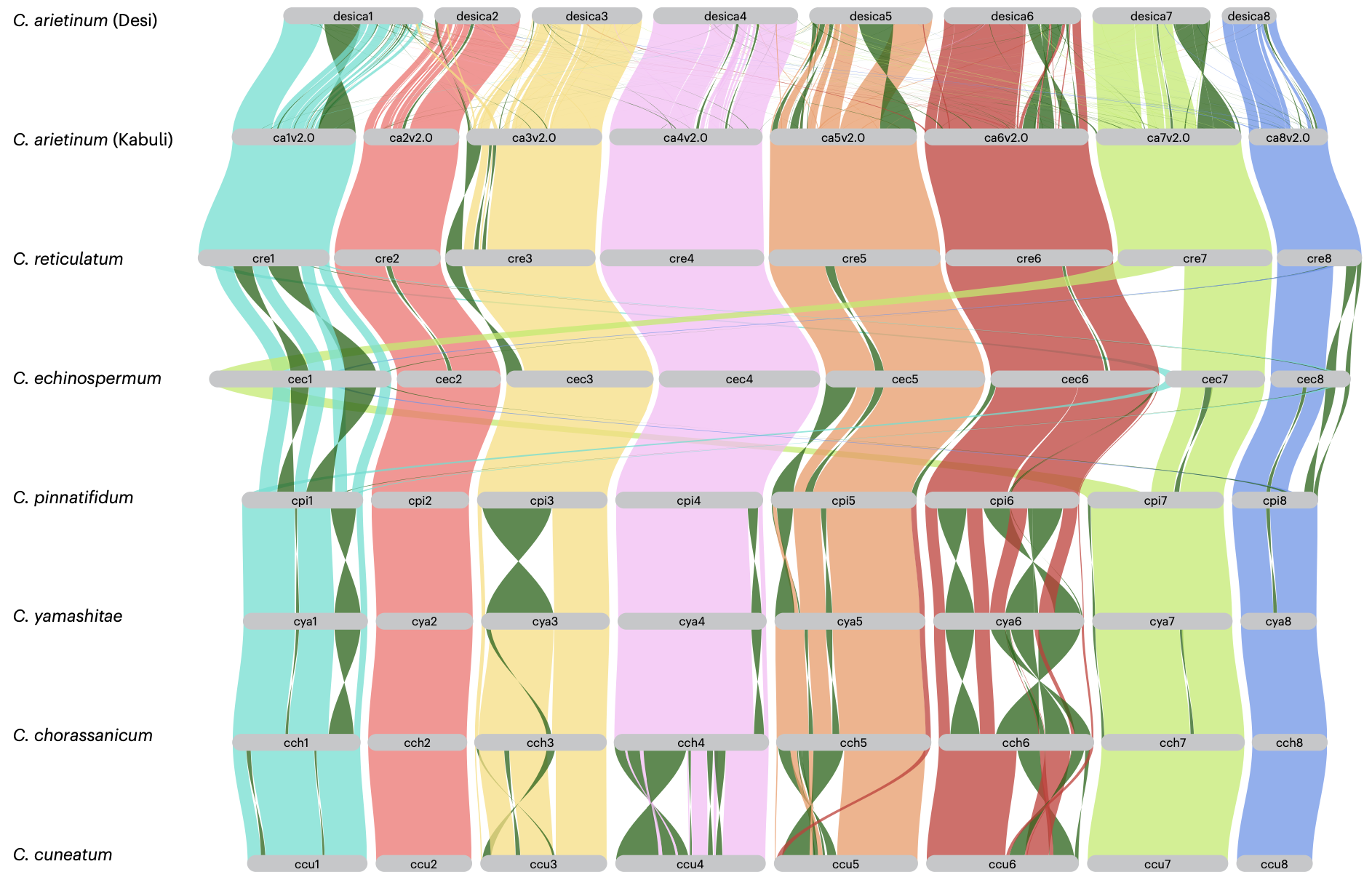
Chickpea, an important legume crop, has limited genetic diversity, which can hinder efforts to improve its resilience and productivity. Addressing this challenge, an international team of researchers, including Xin Liu from BGI Research and collaborators from Murdoch University, ICRISAT, and the University of Western Australia, has investigated the genetic resources within chickpea’s wild relatives. Their research, published in Nature Genetics, describes the development of a “super-pangenome” for the Cicer genus.
The study successfully generated high-quality genome assemblies for eight annual wild Cicer species. This work provides valuable genomic information for these species, which was previously lacking and limited their use in breeding.
Developing the Cicer Super-Pangenome
By analyzing these eight new wild genomes alongside existing cultivated chickpea genomes, the team constructed a Cicer super-pangenome, representing a broad collection of genes found across these species. Key findings include:
- Gene Content: The super-pangenome comprises 24,827 gene families, categorized into core (present in all), softcore, dispensable (present in some), and species-specific groups.
- Agronomic Relevance: The dispensable gene set showed enrichment for genes related to functions like defense responses, potentially offering traits useful for breeding.
- Evolutionary Context: The study provided updated phylogenetic relationships and divergence time estimates for the annual Cicer species.
- Genetic Variation: A comprehensive catalog of genetic variations (SNPs, InDels, and large Structural Variations - SVs) was created, highlighting differences between wild and cultivated species.
- Trait-Associated Variations: Notably, variations, including SVs, were identified in or near genes known to influence traits like flowering time, vernalization, and disease resistance.
Graph Genome Construction
To better utilize the complex variation data, especially SVs, the researchers built a graph-based pangenome. This structure integrates variations into the reference, improving the accuracy of read mapping and SV genotyping across different Cicer accessions compared to using a single linear reference.
Impact and Application
We suggest that this Cicer super-pangenome and the associated graph genome are useful resources for the research community. They provide a deeper understanding of genetic diversity within the genus and identify specific genetic variations in wild relatives that could be relevant for improving cultivated chickpea. This information is expected to facilitate the development of improved chickpea varieties using marker-assisted selection or gene editing techniques.
Our Collaborative Role and Brief Reflections
Together with our long‑term partners led by Dr. Rajeev Varshney, we sequenced and assembled genomes from multiple legume species within the Cicer branch. For the first time, we proposed the concept of a super‑pangenome, and through comparative genomic analysis, we clearly delineated significant genomic differences between wild Cicer species and cultivated chickpea—particularly in the form of numerous structural variations that influence key functional genes. Our collaborators further validated that some of these structural variations directly affect traits such as flowering time.
This study offers a comprehensive look into the genetic diversity and evolutionary history of chickpea. By capturing the full spectrum of wild gene pools and integrating them into a super‑pangenome, our work provides essential information for advancing chickpea breeding and crop improvement. We believe that these genomic resources and insights will serve as a critical reference for researchers and breeders worldwide.
Read the full paper: Khan, A.W., Garg, V., Sun, S. et al. Cicer super-pangenome provides insights into species evolution and agronomic trait loci for crop improvement in chickpea. Nat Genet 56, 1225–1234 (2024). Published online: 23 May 2024. https://doi.org/10.1038/s41588-024-01760-4