Exploring Antarctic Krill Mitogenomes: Insights into Genetic Diversity and Population Dynamics
Antarctic krill (Euphausia superba), a keystone species in the Southern Ocean ecosystem, plays a pivotal role in global marine biodiversity and biogeochemical cycles. In our recent study, we leveraged previously published whole-genome sequencing data of Antarctic krill to conduct a deeper analysis of mitochondrial genomes, shedding light on population evolution and genetic selection pressures.
Key Findings
-
High-Quality Mitogenome Assembly:
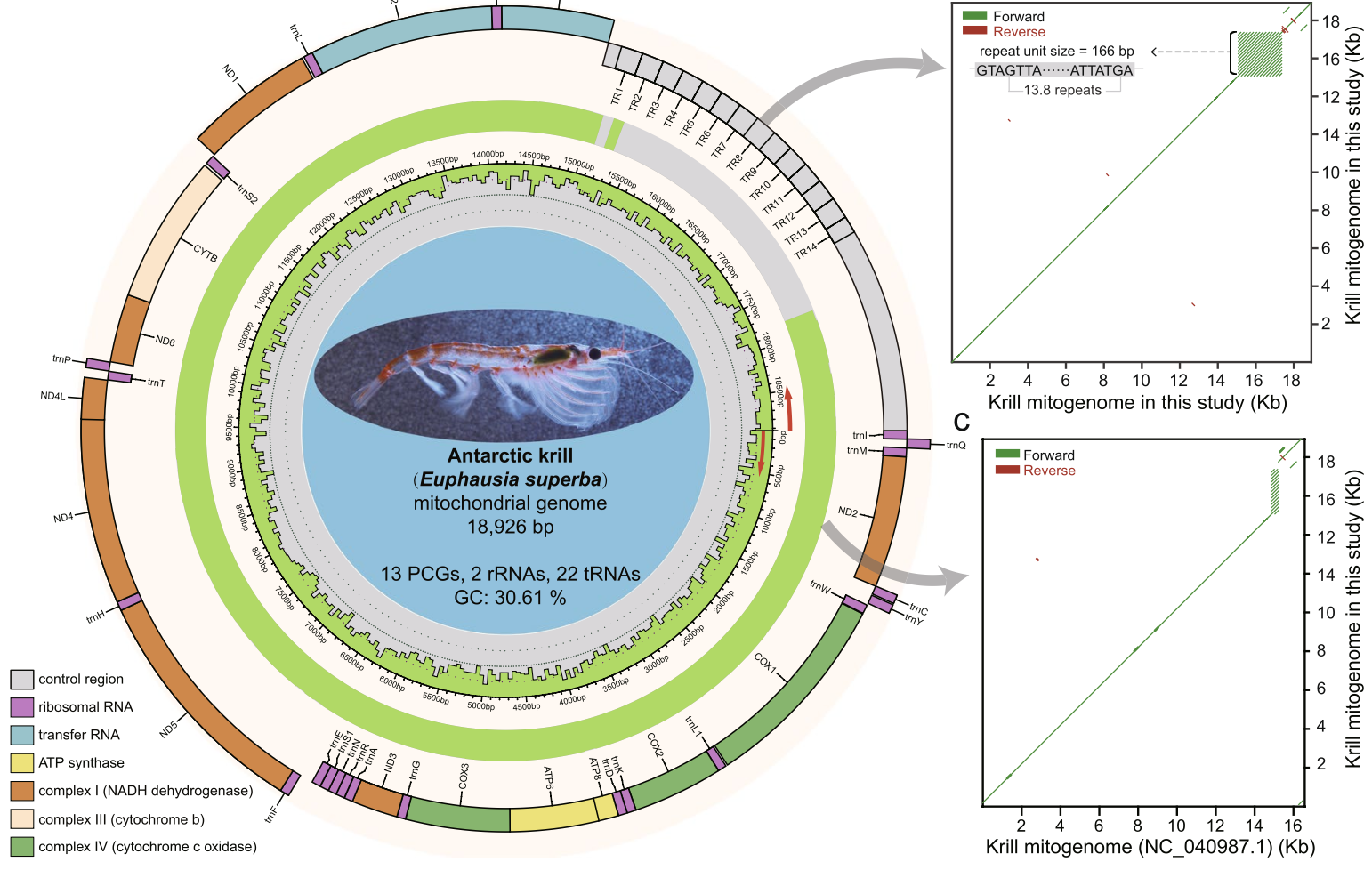
Using PacBio long-read sequencing technology, we assembled the complete mitochondrial genome of Antarctic krill, spanning 18,926 bp with a notably large control region (CR) of 3,952 bp. The CR includes a 2,289 bp tandem repeat, highlighting the advantages of long-read sequencing in resolving complex genomic regions. -
NUMTs and Mitogenome Diversity:
Our analysis identified 900 nuclear-mitochondrial DNA segments (NUMTs) in the krill nuclear genome, totaling 2.79 Mb. These NUMTs provide insights into the dynamic integration of mitochondrial DNA into nuclear genomes during and after speciation events. Additionally, we constructed a dataset of 80 krill mitogenomes, revealing substantial mitochondrial diversity and high levels of genetic connectivity across geographically distinct populations in the Southern Ocean. -
Population Evolution and Demographic History:
Haplotype network analysis and demographic reconstructions suggest a recent population expansion, likely driven by favorable environmental conditions during the late Pleistocene. Our study also revealed evidence of purifying selection across all 13 mitochondrial protein-coding genes, emphasizing the evolutionary constraint to maintain mitochondrial function under extreme conditions.
Reflections and Comments
This project forms part of the PhD research undertaken by Shuai Sun, one of my doctoral students and a vital member of XinLab. Shuai has contributed significantly to various projects involving crop population genomics and marine species, publishing multiple papers in these fields. This particular study represents a focused effort to analyze mitochondrial genomes and uncover evolutionary insights from mutations and selection pressures within krill populations.
As a mentor, I was impressed by Shuai’s dedication and meticulous approach to this research. Leveraging previously published Antarctic krill genomic data, Shuai conducted an in-depth exploration of mitochondrial genes to reveal their evolutionary dynamics and functional significance. These findings not only advance our understanding of krill population biology but also highlight the importance of integrative genomics for marine species conservation.
Conclusion
Our study provides valuable insights into the mitochondrial genomic landscape of Antarctic krill, offering new perspectives on population genetics and evolutionary history. By combining high-quality genomic data with advanced bioinformatics approaches, we have established a foundational resource for krill conservation and management in the Southern Ocean. This work underscores the critical role of mitochondrial research in addressing broader ecological and evolutionary questions.